
RTP File Extension | Gromacs Residue Topology Parameter File | Associated Programs | Free Online Tools - FileProInfo

How to build step wise the topology file for GROMACS (compatible with OPLS force field) for a newly designed ionic liquid?

Data including GROMACS input files for atomistic molecular dynamics simulations of mixed, asymmetric bilayers including molecular topologies, equilibrated structures, and force field for lipids compatible with OPLS-AA parameters - ScienceDirect

TopoGromacs: Automated Topology Conversion from CHARMM to GROMACS within VMD. - Abstract - Europe PMC













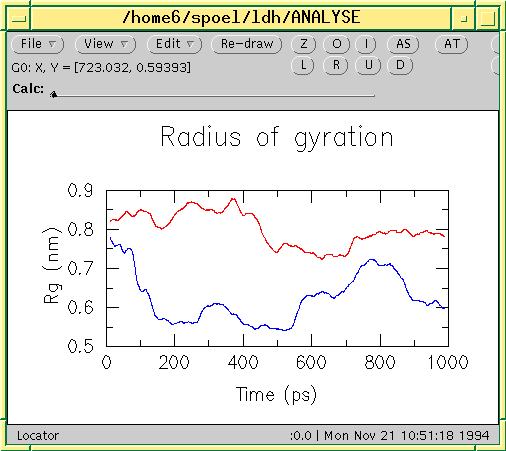

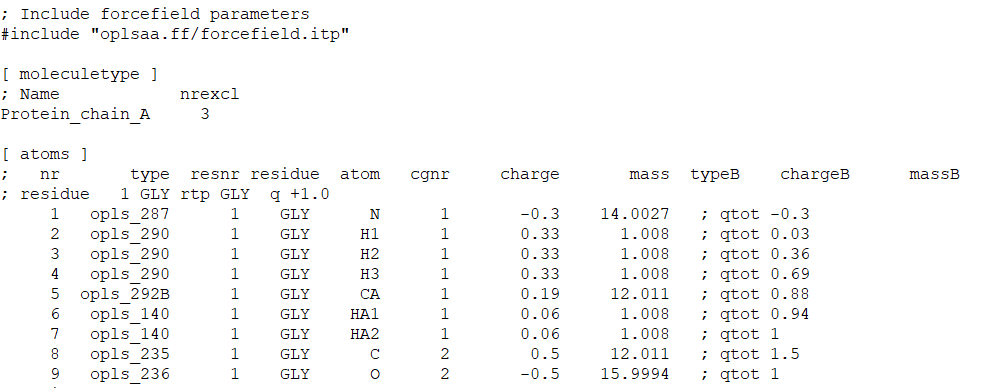

![PDF] GROMACS: A message-passing parallel molecular dynamics implementation | Semantic Scholar PDF] GROMACS: A message-passing parallel molecular dynamics implementation | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/625ddfe164e3a25405672ecc1ca17d05353ff182/8-Table1-1.png)